Counting Polygons in Node Graphs¶
Motivation
- This is what happens when you get curious about efficiently finding triangles in a node graph using linear algebra (instead of recursively walking the whole graph).
Features of custom GraphCounts class:
- Accepts node/edge data in dictionary format
- Plots the graph
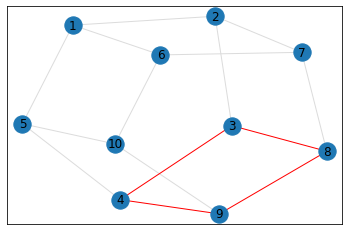
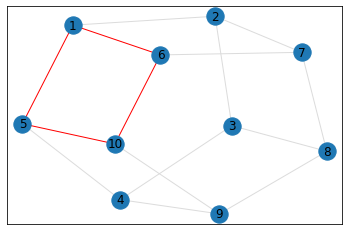
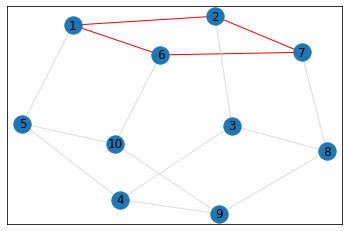
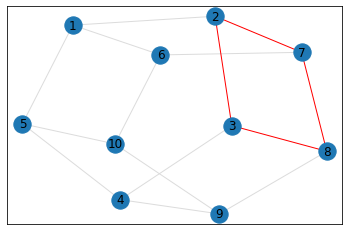
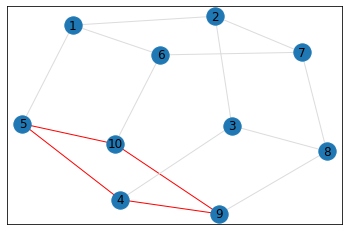
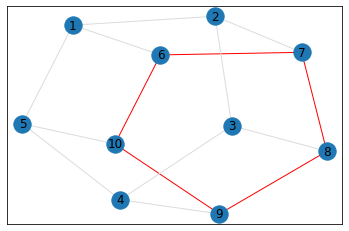
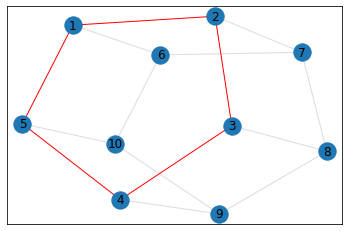
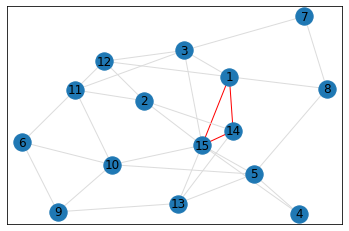
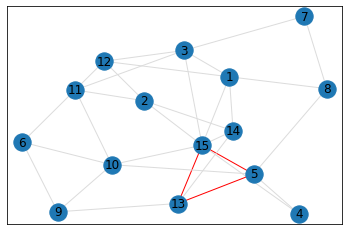
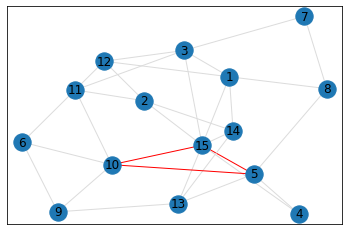
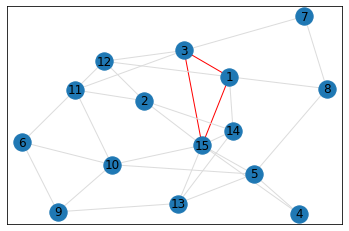
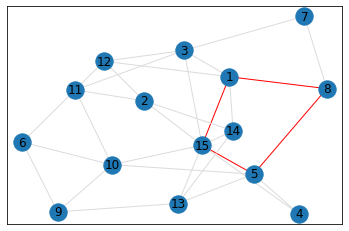
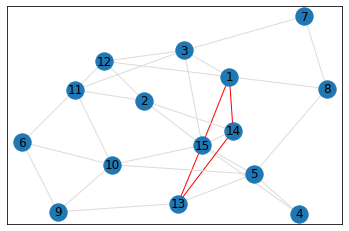
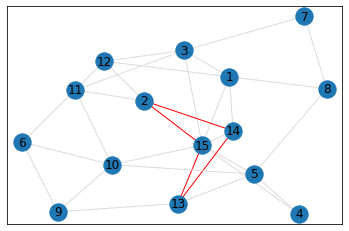
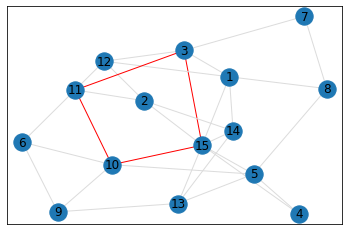
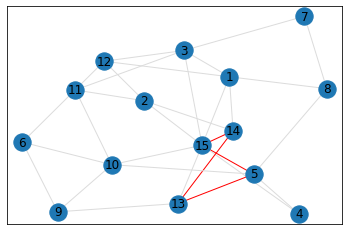
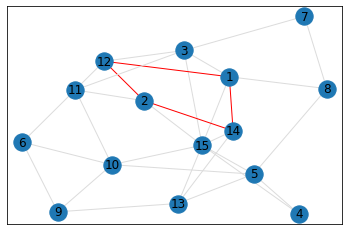
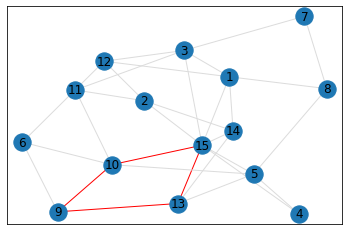
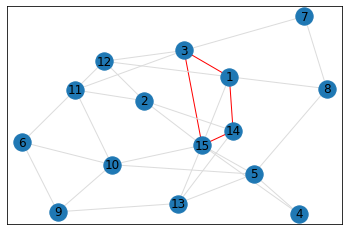
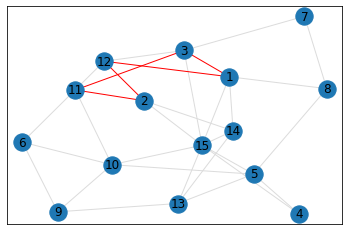
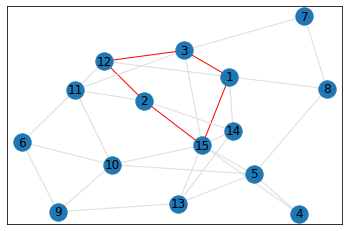
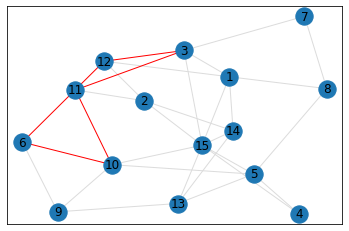
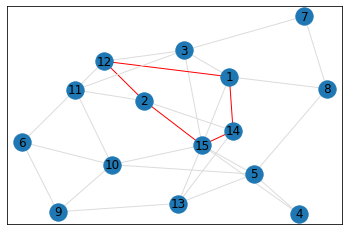
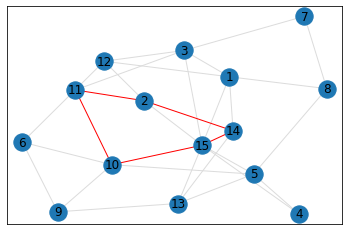
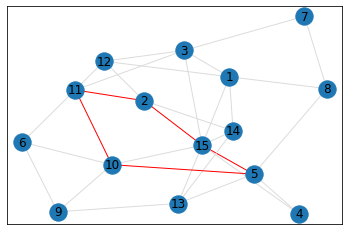
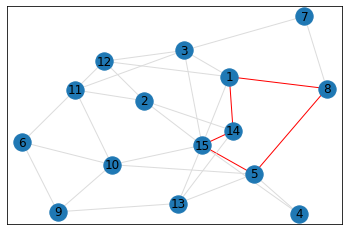
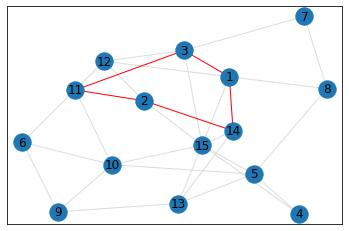
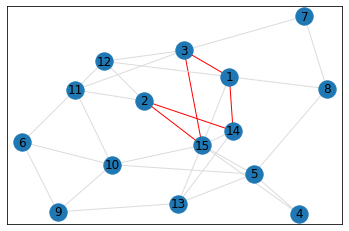
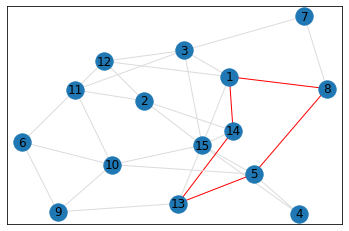
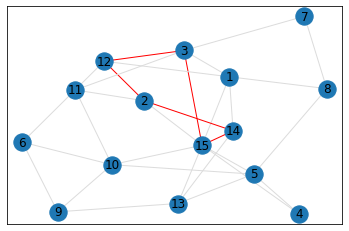
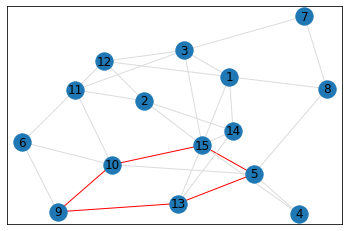
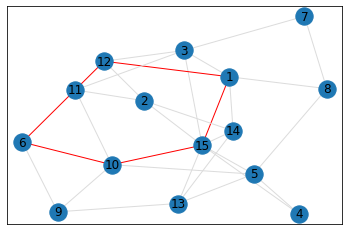
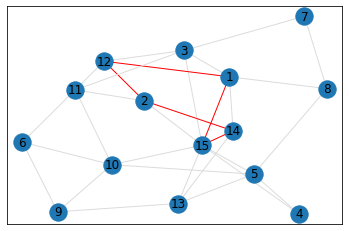
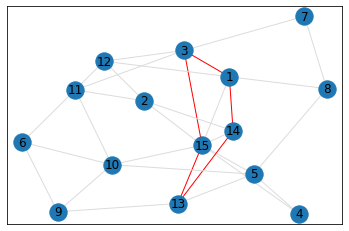
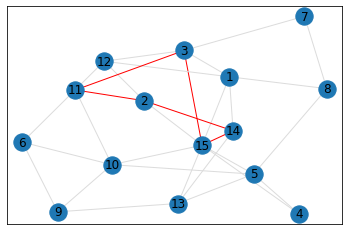
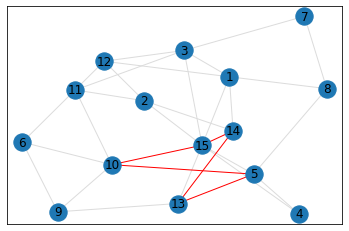
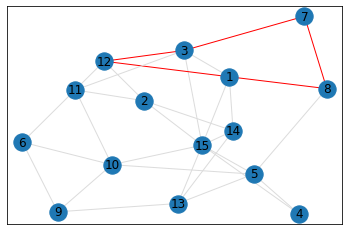
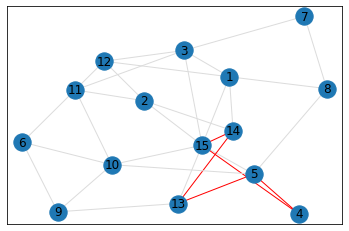
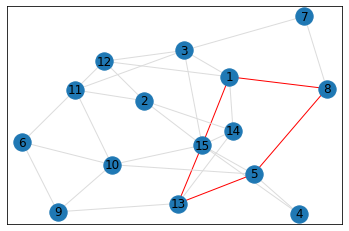
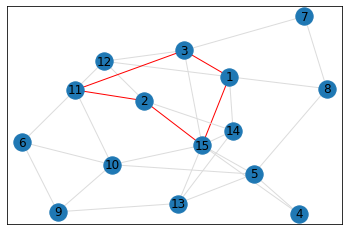
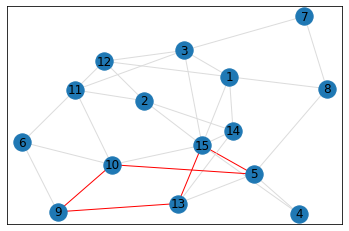
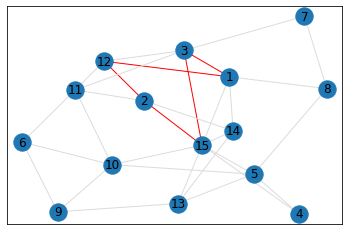
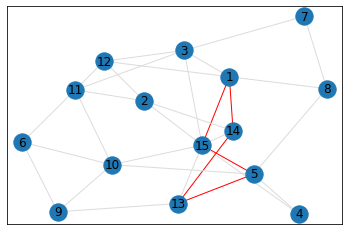
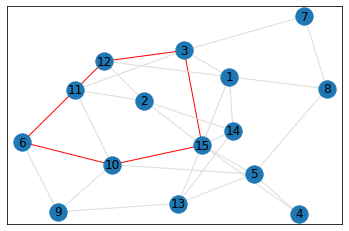
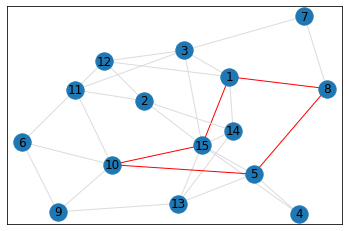
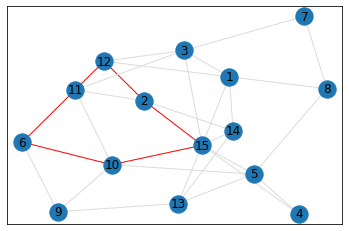
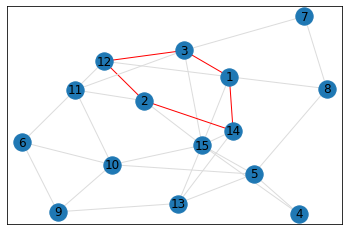
- Counts the number of triangles, quadrilaterals, and pentagons.
- Optionally displays the polygons within the nodes of the graph.
- Written in python (view source code at bottom).
Author:
- Doug Issichopoulos, dougissi.com
- February 10, 2020
Examples¶
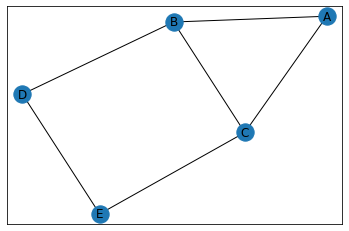
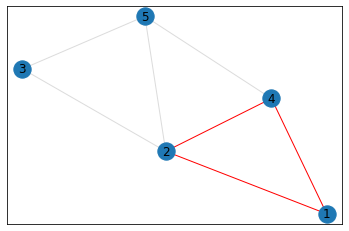
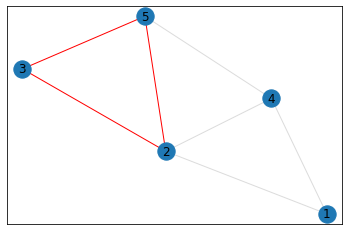
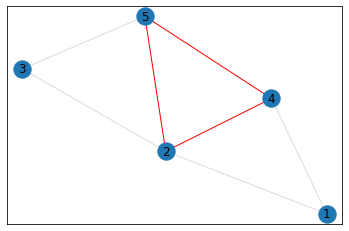
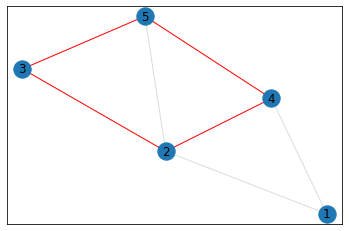
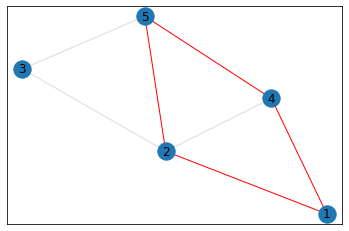
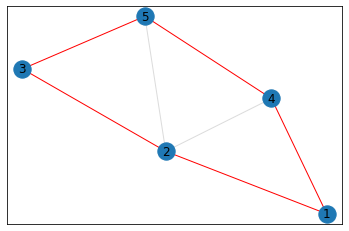
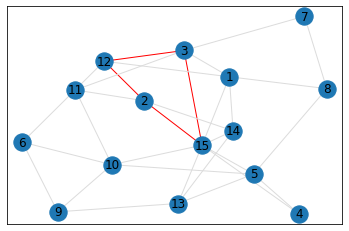
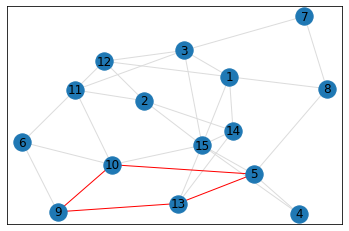
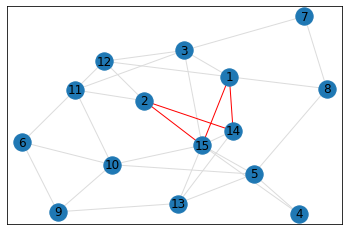
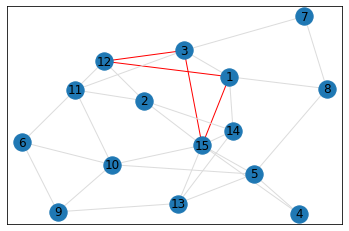
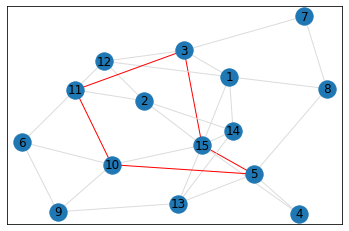
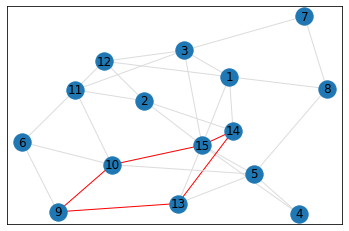
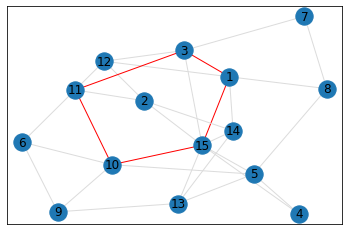
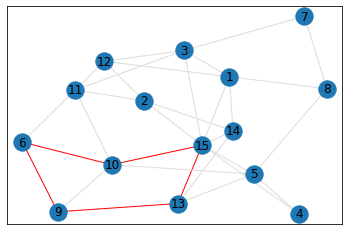
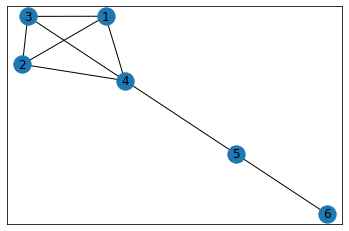
"House"¶
In [2]:
# Graph Data
# `data` maps a node to its set of neighbors
# note: if A has neighbor B, then not needed to specify that B has neighbor A
data = {
'A':{'B', 'C'},
'B':{'C', 'D'},
'E':{'C', 'D'}
}
graph = GraphCounts(data, show_matrix=True, show_graph=True)
In [3]:
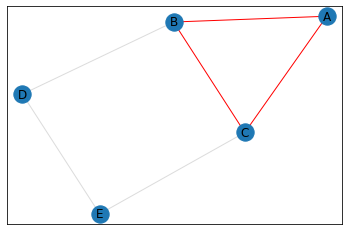
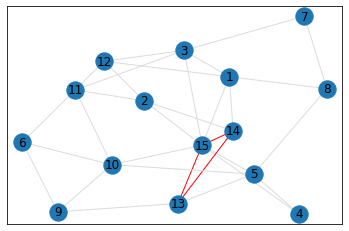
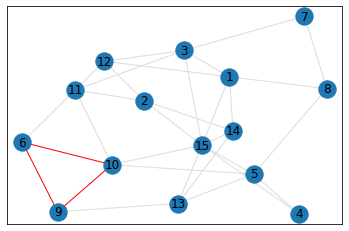
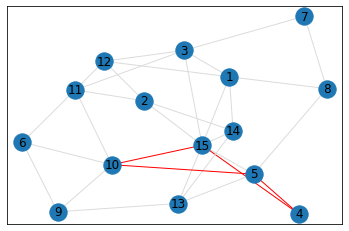
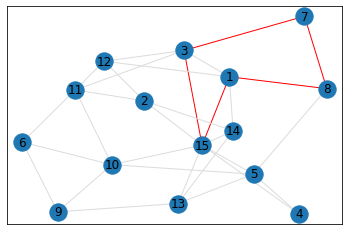
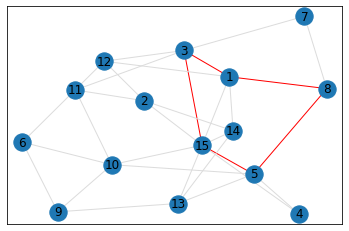
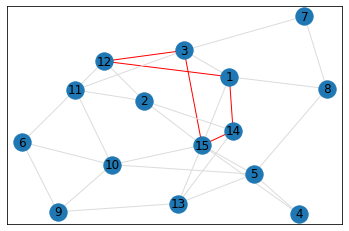
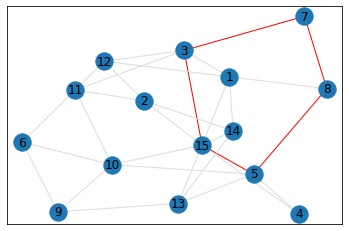
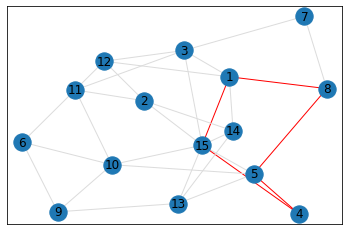
triangle_count = graph.count_triangles(show_plots=True)
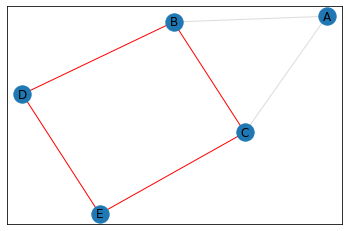
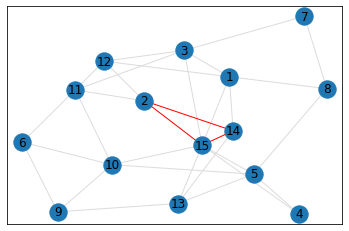
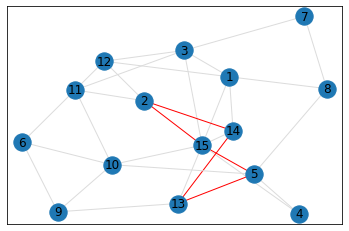
quad_count = graph.count_quads(show_plots=True)
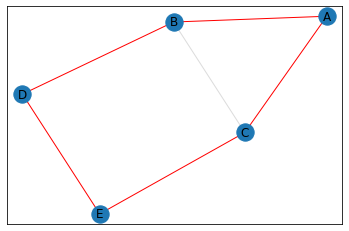
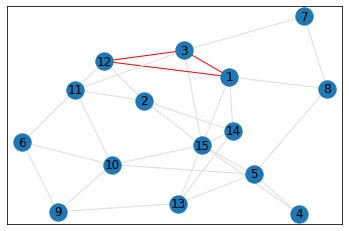
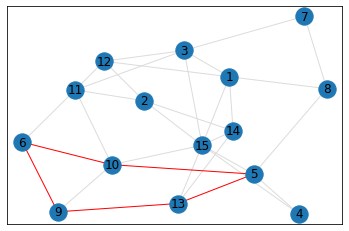
pentagon_count = graph.count_pentagons(show_plots=True)
print("\n\nCounts")
print("triangles count:", triangle_count)
print("quads count:", quad_count)
print("pentagons count:", pentagon_count)
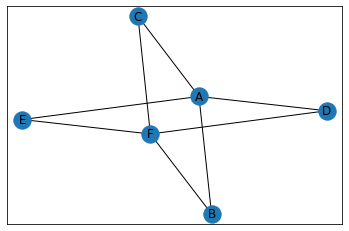
"Trapezoid"¶
In [4]:
data = {
2:{1,3,4,5},
3:{5},
4:{1,5},
}
graph = GraphCounts(data)
In [5]:
triangle_count = graph.count_triangles(show_plots=True)
quad_count = graph.count_quads(show_plots=True)
pentagon_count = graph.count_pentagons(show_plots=True)
print("\n\nCounts")
print("triangles count:", triangle_count)
print("quads count:", quad_count)
print("pentagons count:", pentagon_count)
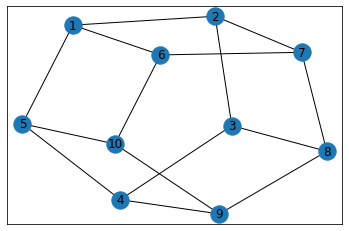
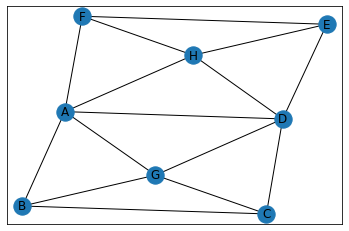
"Pentagonal Prism"¶
In [6]:
data = {
1:{2,5,6},
2:{3,7},
3:{4,8},
4:{5,9},
5:{10},
6:{7,10},
8:{7,9},
9:{10}
}
graph = GraphCounts(data)
In [7]:
triangle_count = graph.count_triangles(show_plots=True)
quad_count = graph.count_quads(show_plots=True)
pentagon_count = graph.count_pentagons(show_plots=True)
print("\n\nCounts")
print("triangles count:", triangle_count)
print("quads count:", quad_count)
print("pentagons count:", pentagon_count)
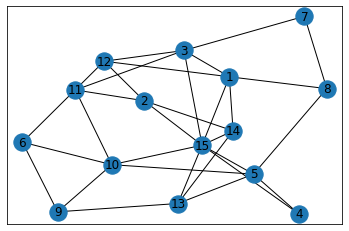
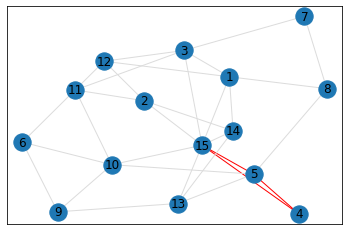
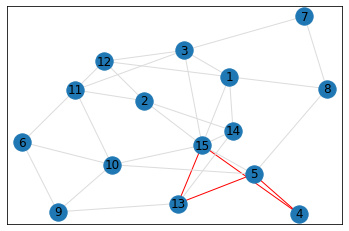
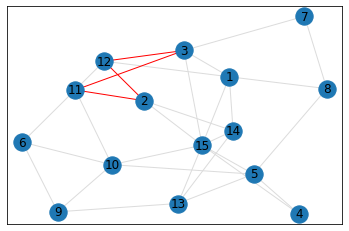
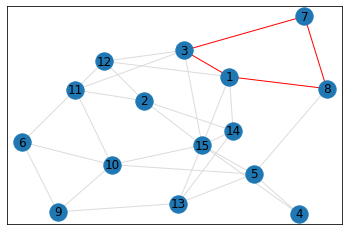
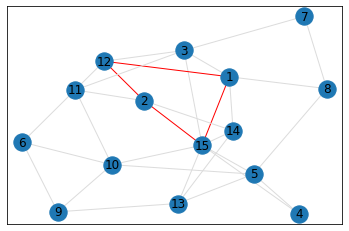
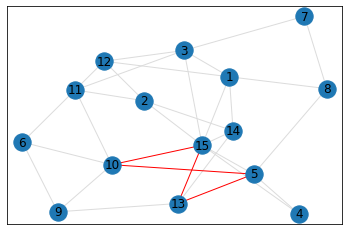
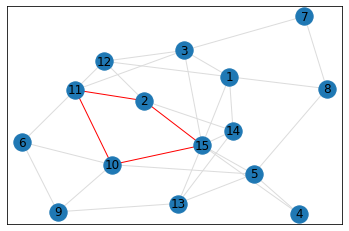
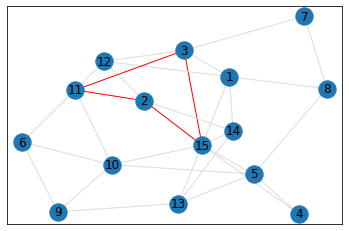
Random, 15 nodes¶
In [8]:
# generate random graph
data = make_random_graph_data(15, 3)
graph = GraphCounts(data)
In [9]:
triangle_count = graph.count_triangles(show_plots=True)
quad_count = graph.count_quads(show_plots=True)
pentagon_count = graph.count_pentagons(show_plots=True)
print("\n\nCounts")
print("triangles count:", triangle_count)
print("quads count:", quad_count)
print("pentagons count:", pentagon_count)
Random, 1,000 nodes¶
A graph this large needs a much larger display, so graphs will not be shown in this example. But note the efficient execution times.
In [10]:
# generate random graph
data = make_random_graph_data(1000, 3)
graph = GraphCounts(data, show_matrix=False, show_graph=False)
# runtime: 356 milliseconds
In [11]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
# runtime: 256 milliseconds
Random, 10,000 nodes¶
A graph this large needs a much larger display, so graphs will not be shown in this example. But note the efficient execution times.
In [20]:
# generate random graph
data = make_random_graph_data(10000, 3)
graph = GraphCounts(data, show_matrix=False, show_graph=False)
# runtime: 21.6 seconds
In [21]:
graph.count_triangles()
# runtime: 63.0 seconds
Out[21]:
In [22]:
graph.count_quads()
# runtime: 57.2 seconds
Out[22]:
In [23]:
graph.count_pentagons()
# runtime: 89.1 seconds
Out[23]:
Other Examples¶
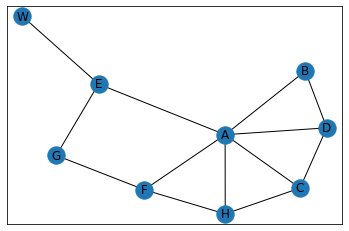
"Kite"¶
In [25]:
data = {
1:{2,3,4},
2:{1,3,4},
3:{1,2,4},
4:{1,2,3},
5:{4,6}
}
graph = GraphCounts(data)
In [28]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
"Ninja Star"¶
In [30]:
data = {
'A': {'B', 'C', 'D', 'E'},
'F': {'B', 'C', 'E', 'D'},
}
graph = GraphCounts(data)
In [31]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
"Two Windows"¶
In [32]:
data = {
'A':{'B', 'G', 'D', 'H', 'F'},
'B':{'C', 'G'},
'C':{'G'},
'D':{'C', 'G', 'H', 'E'},
'E':{'H'},
'F':{'H', 'E'},
}
graph = GraphCounts(data)
In [33]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
"Baby Carriage"¶
In [34]:
# maps a node to its set of neighbors
# note: if A has neighbor B, then not needed to specify that B has neighbor A
# rects with triangles
data = {
'A': {'B','C', 'D', 'E', 'F'},
'D': {'B', 'C'},
'G': {'E', 'F'},
'H': {'A', 'C', 'F'},
'E': {'W'}
}
graph = GraphCounts(data)
In [35]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
"Bipolar"¶
In [36]:
data = {
1:{2,3,10},
3:{2,5},
5:{10},
10:{4},
4:{6,7,8},
7:{6,8,9},
8:{9}
}
graph = GraphCounts(data)
In [37]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
"Sand Dollar"¶
In [39]:
# star
data = {
1:{2,5,6,7,10},
2:{3,6,7,8},
3:{4,7,8,9},
4:{5,8,9,10},
5:{6,9,10},
6:{7,10},
8:{7,9},
9:{10}
}
graph = GraphCounts(data)
In [40]:
print("triangles:", graph.count_triangles())
print("quadrilaterals:", graph.count_quads())
print("pentagons:", graph.count_pentagons())
In [1]:
import pandas as pd
import numpy as np
import networkx as nx
from collections import defaultdict
import random
import matplotlib.pyplot as plt
def symmetrify_dict_of_dicts(dict_of_dicts):
sym_dict = defaultdict(dict, dict_of_dicts)
for k, v in dict_of_dicts.items():
for x, y in v.items():
sym_dict[x][k] = y
return dict(sym_dict)
def symmetrify_dict_of_sets(dict_of_sets):
sym_dict = defaultdict(set, dict_of_sets)
for k, v in dict_of_sets.items():
for x in v:
sym_dict[x].add(k)
return dict(sym_dict)
def matrix_multiplication(x, y):
return np.matmul(x, y)
def matrix_to_power(S, n):
Sn = S # initialize output
while n > 1:
Sn = matrix_multiplication(Sn, S)
n -= 1
return Sn
def get_matrix_diagonal_dict(S, nodes):
'''`nodes` should be ordered the same as the rows/columns of `S`'''
return {node: val for node, val in zip(nodes, S.diagonal())}
def make_random_graph_data(node_count, max_initial_neighbors):
all_nodes = list(range(1, node_count+1))
data = {}
for node in all_nodes:
all_other_nodes = set(all_nodes).difference({node})
rand_num_neighbors = random.randrange(1,max_initial_neighbors+1)
data[node] = set(random.sample(all_other_nodes, rand_num_neighbors))
# print counts of nodes and edges, as well as avg degree, to console
all_edges = set()
degs = []
for node1, edges_set in data.items():
for node2 in edges_set:
all_edges.add(tuple(sorted([node1, node2])))
degs.append(len(edges_set))
print(f'Node count: {len(all_nodes)}, edge_count: {len(all_edges)}, avg degree: {sum(degs)/len(degs)}')
return data
class GraphCounts(object):
def __init__(self, data, show_matrix=False, show_graph=True):
'''
data can be dict from node -> {neighbor set}, or node -> {neighbor1: weight, neighbor2: weight,...},
where if B is a neighbor of A, no need to define A as neighbor of B, as weights are ignored.
'''
self.data_dict = symmetrify_dict_of_dicts({k:{x:1 for x in list(v) if x != k} for k, v in data.items()})
# make pandas DataFrame of graph data
df = pd.DataFrame(self.data_dict).fillna(0)
df.sort_index(inplace=True)
df.sort_index(axis=1, inplace=True)
self.df = df
# make matrix
self.S1 = df.values
if show_matrix:
print('S1:\n',self.S1)
# optionally plot graph data using networkx
if show_graph:
self.G, self.pos = self._define_graph()
nx.draw_networkx(self.G, pos=self.pos)
plt.show()
def _define_graph(self):
G = nx.Graph(self.df)
pos = nx.spring_layout(G)
return G, pos
def _get_triangles_dict(self, S3):
returning_3_walks_dict = get_matrix_diagonal_dict(S3, self.df.index)
return {k: v / 2 for k, v in returning_3_walks_dict.items()}
@staticmethod
def _get_nodes_set_with_unidentified_polygons(polygons_dict):
return {node for node, polygon_count in polygons_dict.items() if polygon_count > 0}
@staticmethod
def _polygon_data_to_tuple(data):
'''
data should be dict from node -> {neighbor set},
where if B is a neighbor of A, no need to define A as neighbor of B
'''
sym_dict = symmetrify_dict_of_sets(data)
nodes = set()
edges = set()
for node, edges_set in sym_dict.items():
nodes.add(node)
for edge in edges_set:
edges.add(tuple(sorted({node, edge})))
return (tuple(sorted(nodes)), tuple(sorted(edges)))
def _identify_polygons(self, polygon_type, polygons_dict):
# helper function
def build_polygons_set(simplified_data_dict, polygon_type, polygons_set, polygons_dict):
'''
Builds `polygons_set` via recursion, based on number of polygons calculated at each node (from `polygons_dict`)
'''
for node1, node1_neighbors in simplified_data_dict.items():
for node2 in list(node1_neighbors):
node2_neighbors = simplified_data_dict[node2]
other_node1_neighbors = node1_neighbors.difference({node2})
other_node2_neighbors = node2_neighbors.difference({node1})
if polygon_type == 'triangle':
triangles = set()
common_neighbors = other_node1_neighbors.intersection(other_node2_neighbors)
for node3 in list(common_neighbors):
triangle = self._polygon_data_to_tuple({
node1: {node2, node3},
node2: {node3}
})
if triangle not in polygons_set:
triangles.add(triangle)
polygons = triangles
elif polygon_type == 'quad':
connections = set()
quads = set()
for node3 in list(other_node1_neighbors):
for node4 in list(other_node2_neighbors):
if node3 != node4 and node4 in simplified_data_dict[node3]: # nodes different and connection exists between them
connection = tuple(sorted([node3, node4]))
if connection not in connections: # if new connection then valid polygon
connections.add(connection)
quad = self._polygon_data_to_tuple({
node1: {node2, node3},
node2: {node4},
node3: {node4}
})
if quad not in polygons_set:
quads.add(quad)
polygons = quads
elif polygon_type == 'pentagon':
connections = set()
pentagons = set()
for node3 in list(other_node1_neighbors):
for node4 in list(other_node2_neighbors):
if node3 != node4: # nodes different
common_neighbors = set(simplified_data_dict[node3]).intersection(simplified_data_dict[node4]).difference({node1,node2,node3,node4})
for node5 in common_neighbors:
connection = tuple(sorted([node3, node4, node5]))
if connection not in connections: # if new connection then valid polygon
connections.add(connection)
pentagon = self._polygon_data_to_tuple({
node1: {node2, node3},
node2: {node4},
node3: {node5},
node4: {node5}
})
if pentagon not in polygons_set:
pentagons.add(pentagon)
polygons = pentagons
else:
raise ValueError(f"Polygon type '{polygon_type}' not valid.")
if not polygons:
continue
# update storages based on new polygons
for polygon in polygons:
# add polygon to storage
polygons_set.add(polygon)
# decrement count of unidentified polygons at node, for each node in polygon
nodes, _ = polygon
for node in nodes:
polygons_dict[node] -= 1
return
## main function
# raise error if invalid polygon type
valid_polygon_types = {'triangle', 'quad', 'pentagon'}
if polygon_type not in valid_polygon_types:
raise ValueError(f"Polygon type '{polygon_type}' not valid -- must be among: {valid_polygon_types}")
polygons_dict = dict(polygons_dict) # make copy to avoid mutilation
nodes_set_with_unidentified_polygons = self._get_nodes_set_with_unidentified_polygons(polygons_dict)
simplified_data_dict = {k:{x for x, _ in v.items() if x in nodes_set_with_unidentified_polygons}
for k, v in self.data_dict.items()
if k in nodes_set_with_unidentified_polygons}
polygons_set = set()
while True:
build_polygons_set(simplified_data_dict, polygon_type, polygons_set, polygons_dict)
# limit search based on found polygons above
nodes_set_with_unidentified_polygons = self._get_nodes_set_with_unidentified_polygons(polygons_dict)
if not nodes_set_with_unidentified_polygons:
break
simplified_data_dict = {k:{x for x in v if x in nodes_set_with_unidentified_polygons}
for k, v in simplified_data_dict.items()
if k in nodes_set_with_unidentified_polygons}
return polygons_set
def _plot_polygons(self, polygons_set, desc):
def show_original_graph(G, pos):
print('Original')
nx.draw_networkx(G, pos=pos)
plt.show()
# if graph wasn't originally drawn, define it
if not hasattr(self, 'G') or not hasattr(self, 'pos'):
self.G, self.pos = self._define_graph()
if not polygons_set: # if no polygons to plot, show original anyway
show_original_graph(self.G, self.pos)
# if no polygons to plot, inform user and return
if not polygons_set:
print(f'{desc}: none to show')
return
all_graph_edges = [tuple(sorted(edge)) for edge in self.G.edges]
# plot each polygon
print(desc)
for _, edges in polygons_set:
polygon_edges = set(edges)
edge_colors = ["red" if edge in polygon_edges else "gainsboro" for edge in all_graph_edges]
nx.draw_networkx(self.G, pos=self.pos, edge_color=edge_colors)
plt.show()
def count_triangles(self, show_plots=False):
S3 = matrix_to_power(self.S1, 3)
if show_plots:
triangles_dict = self._get_triangles_dict(S3)
triangles_set = self._identify_polygons('triangle', triangles_dict)
self._plot_polygons(triangles_set, 'Triangles')
return np.trace(S3) / 6
def count_quads(self, show_plots=False):
S2 = matrix_to_power(self.S1, 2)
S4 = matrix_to_power(S2, 2)
degrees_dict = get_matrix_diagonal_dict(S2, self.df.index)
returning_4_walks_dict = get_matrix_diagonal_dict(S4, self.df.index)
# make dict mapping a node to the count of all quadrilaterals passing it
quads_dict = {}
for node in self.data_dict.keys():
quads_dict[node] = 0 # assume none unless proven otherwise
returning_4_walks_at_node = returning_4_walks_dict[node]
if returning_4_walks_at_node < 8:
continue
deg = degrees_dict[node]
if deg < 2:
continue
sum_neighbor_degs = sum([degrees_dict[neighbor] for neighbor in self.data_dict[node]])
if sum_neighbor_degs < 4:
continue
quads_dict[node] = (returning_4_walks_at_node - (deg * (deg - 1) + sum_neighbor_degs)) / 2
if show_plots:
quads_set = self._identify_polygons('quad', quads_dict)
self._plot_polygons(quads_set, 'Quadrilaterals')
return sum(quads_dict.values()) / 4 # account for double counting for each of quad's 4 nodes
def count_pentagons(self, show_plots=False):
S2 = matrix_to_power(self.S1, 2)
S3 = matrix_multiplication(S2, self.S1)
S5 = matrix_multiplication(S3, S2)
returning_5_walks_dict = get_matrix_diagonal_dict(S5, self.df.index)
# get list of triangles
triangles_dict = self._get_triangles_dict(S3)
triangles = self._identify_polygons('triangle', triangles_dict)
# make map from node to list of triangles at that node
triangles_at_nodes_dict = {node: [] for node in self.data_dict}
for triangle in triangles:
nodes, _ = triangle
for node in nodes:
triangles_at_nodes_dict[node].append(nodes)
pentagons_dict = {}
for node in self.data_dict.keys():
pentagons_dict[node] = 0 # assume none unless proven otherwise
returning_5_walks_dict_at_node = returning_5_walks_dict[node]
if returning_5_walks_dict_at_node < 2:
continue
triangles_at_node = triangles_at_nodes_dict[node]
# walks from triangles at node
walks_from_triangles_at_node = 10 * len(triangles_at_node)
# walks from all edges adjacent to triangles at node
walks_from_edges_adj_to_triangles_at_node = 0 # initialize
for triangle in triangles_at_node:
# walks from edges from node not part of triangle
tri_neighbors = set(triangle).difference({node})
other_node_neighbors = set(self.data_dict[node]).difference(tri_neighbors)
walks_from_edges_at_node_adj_to_triangle_at_node = 4 * len(other_node_neighbors)
# walks from edges leaving triangle (not at node)
tri_1, tri_2 = tri_neighbors
other_neighbors_tri_1 = set(self.data_dict[tri_1]).difference({node, tri_2})
other_neighbors_tri_2 = set(self.data_dict[tri_2]).difference({node, tri_1})
walks_from_edges_NOT_at_node_adj_to_triangle_at_node = 2 * (len(other_neighbors_tri_1)
+ len(other_neighbors_tri_2))
walks_from_edges_adj_to_triangles_at_node += (walks_from_edges_at_node_adj_to_triangle_at_node
+ walks_from_edges_NOT_at_node_adj_to_triangle_at_node)
# walks from single edges connecting node to other triangles
walks_from_single_edges_connecting_node_to_other_triangles = 0
for neighbor in self.data_dict[node]:
disjoint_neighbor_triangles = [tri for tri in triangles_at_nodes_dict[neighbor] if node not in tri]
walks_from_single_edges_connecting_node_to_other_triangles += (2 * len(disjoint_neighbor_triangles))
# formula
pentagons_dict[node] = (returning_5_walks_dict_at_node
- (walks_from_triangles_at_node
+ walks_from_edges_adj_to_triangles_at_node
+ walks_from_single_edges_connecting_node_to_other_triangles)
) / 2
if show_plots:
pentagons_set = self._identify_polygons('pentagon', pentagons_dict)
self._plot_polygons(pentagons_set, 'Pentagons')
return sum(pentagons_dict.values()) / 5 # account for double counting for each of pentagon's 5 nodes